For several bacterial species, available whole-genome DNA sequences number in the tens or hundreds of thousands. Such datasets allow reconstruction of evolutionary history that includes thousands of sequence changes that have occurred fairly recently. The inferred changes provide information about both the rates of various types of mutation and the forces of natural selection. The study of recent sequence changes, made possible by the availability of sequences of very closely-related isolates, provides an unusual window into natural selection. Purifying selection, which eliminates sufficiently harmful mutations in the long run, has had little time to act, so that only highly deleterious changes are appreciably affected. Furthermore, sequence changes that confer an advantage under rare conditions, but are disfavored and eliminated in the long run, can be identified.
Projects
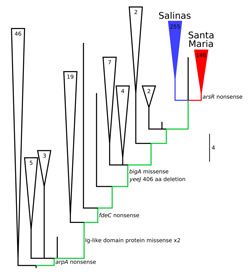
Selection-Driven Gene Inactivation
For a protein-coding gene to persist, selection must favor a functional protein in the long term. Nevertheless, premature stop codons in some genes appear to be positively selected under some circumstances. The identities of affected genes shed light on both the evolutionary phenomenon and the genes themselves.
Determinants of Position-Specific Mutation Rates
Nucleotide positions in a genome exhibit different rates of mutation. Previous work on this project characterized the high C→T mutation rate at 5-methyl C residues in bacteria, and used this to infer methylation patterns. Other work revealed that N4 methylation of C by a particular Type III restriction-modification system is associated with extremely high C→A mutation rate. Additional hypermutation phenomena are under study.
Short-Term Effects of Selection on Coding Sequences
The evolution of coding sequences and their protein products is most commonly studied by comparison of sequences from different species, with divergence times of millions of years. Comparison of sequences from tight genetic clusters of bacteria, with divergence times of years to decades, provides a very different perspective on protein evolution.
Evolution of Pathogen Characteristics
Genetic differences within a bacterial species can have phenotypic consequences that are relevant to disease. Reconstruction of genetic changes that have occurred recently can elucidate the evolution of disease-causing and other important traits on a relatively short time scale (years or decades).
Publications
-
Recent Genetic Changes Affecting Enterohemorrhagic Escherichia coli Causing Recurrent Outbreaks, featuring Fogarty's Joshua L. Cherry as lead author
Microbiology Spectrum, April 25, 2022 -
Extreme C-to-A Hypermutation at a Site of Cytosine-N4 Methylation, featuring Fogarty's Joshua L. Cherry as lead author
mBio, April 13, 2021 -
Selection-Driven Gene Inactivation in Salmonella, featuring Fogarty's Joshua L. Cherry as lead author
Genome Biology and Evolution, February 11, 2020 -
Methylation-Induced Hypermutation in Natural Populations of Bacteria, featuring Fogarty's Joshua L. Cherry as lead author
Journal of Bacteriology, November 26, 2018 -
A practical exact maximum compatibility algorithm for reconstruction of recent evolutionary history, featuring Fogarty's Joshua L. Cherry as lead author
BMC Bioinformatics, February 23, 2017
Contact
Members
- Joshua L. Cherry
- Nídia S. Trovão