Research Highlights: Influenza, dengue & other matters
November/December 2025 | Volume 24 Number 6
.jpg) Photo courtesy of NIAIDSwine flu virus particles (green) attach to and bud from the surface of a cell (purple)
Photo courtesy of NIAIDSwine flu virus particles (green) attach to and bud from the surface of a cell (purple)
How well did the 2023 flu vaccine work?
Influenza, or the flu, is a common virus that affects young and old alike and causes illness that ranges from mild to severe (requiring hospitalization). The virus changes frequently, so vaccines must be updated each year. This study looked at how well the 2023 southern hemisphere flu vaccine protected people across eight countries, using data from 520 hospitals. Results showed the vaccine reduced hospital admissions by about half; it worked especially well in young children and somewhat less effectively in older adults. Protection against severe disease requiring intensive care was even higher. Vaccine effectiveness varied between countries, likely due to differences in flu strains, timing of vaccination, population health, and vaccine types. The authors, Fogarty’s former director Kathy Neuzil, PhD, and Cécile Viboud, PhD, state that large multi-country studies, including their own, help scientists develop better vaccines.
Article:
Harnessing the power of multicountry networks for influenza vaccine monitoring.
Publication:
The Lancet Global Health, February 2025
Tracking flu viruses in Southeastern Asia
Southeastern Asia plays a key role in spreading seasonal flu around the world. Researchers studied flu virus movement in this region from 2007 to 2023, including during the 2009 H1N1 and COVID-19 pandemics. The study showed that COVID-19 greatly disrupted flu spread, stopping the usual waves of influenza virus movement; the 2009 H1N1 pandemic had a smaller effect. The A/H3N2 flu virus persisted more in the region than the B/Victoria virus, while patterns of virus evolution changed depending on the pandemic. Human travel, immunity, and behavior during pandemics affect flu circulation; understanding these patterns helps improve surveillance, vaccine planning, and preparation for future pandemics, noted the authors, including Fogarty’s Cécile Viboud, PhD.
Article:
Disruption of seasonal influenza circulation and evolution during the 2009 H1N1 and COVID-19 pandemics in Southeastern Asia.
Publication:
Nature Communications, January 2025
Data from Denmark reveals hospitalizations, deaths linked to respiratory viruses
A study in Denmark focused on three respiratory viruses—RSV, influenza, and COVID-19—and their impact on adults from 2015 to 2024. All three viruses caused significant deaths and hospitalizations, especially among people aged 65 and older. RSV, often overlooked in adults, caused nearly as many hospital admissions as influenza in older adults, with post-pandemic seasons seeing higher RSV activity. Influenza and COVID-19 also caused serious illness, though widespread vaccination helped reduce this. While COVID-19 lockdowns temporarily lowered the spread of all respiratory viruses, RSV and influenza rebounded after restrictions lifted. Fogarty’s Cécile Viboud, PhD, Chelsea Hansen, PhD, and their co-authors conclude that enhanced RSV testing, monitoring of respiratory viruses, and ongoing vaccination is needed.
Article:
Excess mortality and hospitalisations associated with respiratory syncytial virus, influenza, and COVID-19 among adults in Denmark (2015-2024): a modelling study.
Publication:
The Lancet Regional Health: Europe, August 2025
The power (and limits) of using genomes to track outbreaks
Scientists use pathogen genome sequences to study how diseases spread between groups of people or animals. These sequences change over time as the pathogen mutates; the speed of mutation compared with the speed of disease transmission affects how much scientists can learn. Fast-mutating pathogens can reveal more detailed movement patterns, while slow-mutating ones may show only broad trends. The number of samples also matters—more samples give a clearer picture of how a disease spreads. This study introduces a framework to understand the limits of what genome data can reveal, while strategies like grouping populations, analyzing longer genome segments, or using deep sequencing may improve accuracy. Fogarty’s Amanda Perofsky contributed to this work.
Article:
Characterizing the informativeness of pathogen genome sequence datasets about transmission between population groups.
Publication:
medRxiv(preprint), August 2025
Why tracking MERS requires multiple methods
This study looked at transmission of the Middle East Respiratory Syndrome (MERS) virus and how well different scientific tools can track that movement. Researchers analyzed more than 600 virus genomes collected from 2012 to 2024. All methods confirmed that MERS mainly spreads from infected dromedary camels to humans, mostly in Saudi Arabia and the United Arab Emirates. However, the tools did not always agree on how often these “spillover” events happened—some estimated as few as 15 and others up to 34. The researchers recommend using fast tools for quick outbreak detection and a multi-method review for long-term planning. Continued genomic monitoring of both camels and humans is essential for catching new MERS threats early. Fogarty’s Nidia Trovão, PhD, contributed to this article.
Article:
A scalable maximum-likelihood framework for near-real-time monitoring of MERS-CoV evolutionary and zoonotic dynamics.
Publication:
Microbiology Spectrum, November 2025
Dengue in Saudi Arabia: Tracking a virus across borders
A recent study of dengue virus (DENV) in Saudi Arabia analyzed 20 full virus genomes collected between 2021 and 2023. Researchers found three types of dengue co-circulating there—DENV-1, DENV-2, and DENV-3—the most common being DENV-2. The viruses were repeatedly introduced from countries in South and Southeast Asia and East Africa. Some strains had been circulating in Saudi Arabia undetected for years, showing both ongoing local spread and the country’s role as a hub for regional transmission. Mass gatherings, international travel, and labor migration increased the risk of dengue spread. The study highlights the urgent need for better virus monitoring, stronger mosquito control, and regional cooperation to track outbreaks and reduce disease impact. Fogarty’s Nidia Trovão, PhD, co-authored this study.
Article:
Molecular evolutionary insights into the repeated introductions and cryptic transmission of dengue virus in Saudi Arabia.
Publication:
The Journal of Infectious Diseases, September 2025
Predicting how viruses outsmart immunity
Viruses change most rapidly when immunity is neither too weak nor too strong—this is called the “phylodynamic curve.” Here, Fogarty’s Cécile Viboud, PhD, and co-authors develop a new framework to better understand how viruses evolve to escape immunity in a population. The framework shows how factors like partial immunity, vaccination, seasonal outbreaks, travel, and public health measures affect the risk and timing of the emergence of new virus variants. For example, partially protective immunity and the lifting of interventions (such as mask mandates) can influence when and where immune-evading variants appear. By understanding these dynamics, public health officials can better prevent outbreaks of dangerous viral variants, improve vaccine strategies, and reduce the risk of immune escape.
Article:
Eco-evolutionary dynamics of pathogen immune-escape: deriving a population-level phylodynamic curve.
Publication:
Journal of the Royal Society Interface, April 2025
Floods, dust storms & rising lung disease across the U.S.
Nontuberculous mycobacteria (NTM) are bacteria found in soil and water that can cause long-lasting lung infections, especially in people with lung problems or cystic fibrosis. This study looked at NTM cases across the U.S. and compared them with weather patterns and severe events like floods and dust storms. The researchers found that NTM infections have been rising and that certain weather conditions—such as high temperatures, heavy rain, cloud cover, and changes in air pressure—are linked to higher infection rates. Floods in the Southeast and dust storms in mid-latitude regions were especially strong predictors. The timing between weather events and later increases in infections varied by region. The authors, including Fogarty’s Samantha Bents, highlight the need for better testing, reporting, and awareness.
Article:
Factors predicting incidence of nontuberculous mycobacteria in an era of climate change and altered ecosystems in the United States.
Publication:
The Science of the Total Environment, October 2025
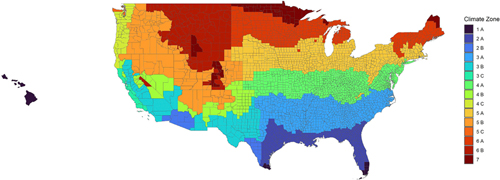 Photo courtesy of the journal The Science of the Total EnvironmentThis colored map segments the U.S. into 15 unique zones based on temperature and moisture levels.
Photo courtesy of the journal The Science of the Total EnvironmentThis colored map segments the U.S. into 15 unique zones based on temperature and moisture levels.
Can data science improve health?
The Data Science for Health Discovery and Innovation in Africa (DS-I Africa) initiative, funded by NIH, includes 38 projects that use tools like artificial intelligence, big data, and genomics to address major health challenges such as cancer, malaria, and air pollution. Led mainly by African researchers, the program brings together universities, governments, nonprofits, and private companies to share data, train new scientists, and create new technologies tailored to African communities. To support the building of strong partnerships, DS-I Africa runs “Networking Exchange” events where people from many countries and fields of study can meet and form collaborations. As the program nears the end of its first funding phase in 2026, it is planning for long-term sustainability. Fogarty’s Laura Povlich, PhD, Amit Mistry, PhD, and Gifty Dankyi contributed to this report.
Article:
How the DS-I Africa Consortium Is Harnessing the Power of Partnerships for Data Science in Africa .
Publication:
Data Science Journal, November 2025
Updated December 12, 2025
To view Adobe PDF files,
download current, free accessible plug-ins from Adobe's website.